More Information
Here, you’ll find more information about the paper and models. Detailed information about the code and how to use it is in the GitHub repository. And requirements for the equipment and input data are explained in Requirements.
Paper’s Results
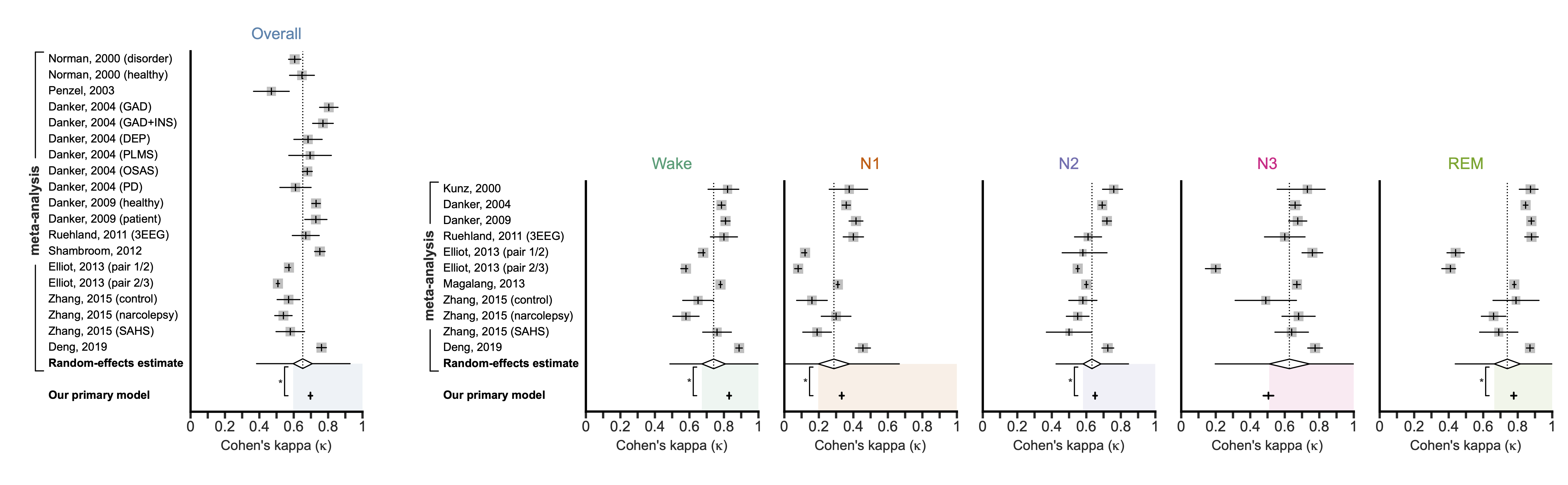
Meta-analysis comparison of PSG- and CSG-based sleep staging
The main takeaway from the paper is that our model performs equivalently to human-scored PSG. To make this comparison, we performed a meta-analysis using the data from 11 human-to-human studies to get the overall and stage-specific kappas that can be expected for humans (i.e., the random-effects estimates). Then, we compared our results against the human performance using non-inferiority testing.
The paper provides more details, including: breakdowns by source study, comparison with EEG-less studies, robustness to noise and other perturbations, and the real-time performance.
Data and Rigor
Below are two important details about the diversity of the data and the methodological rigor. However, the paper provides more details.
Data diversity
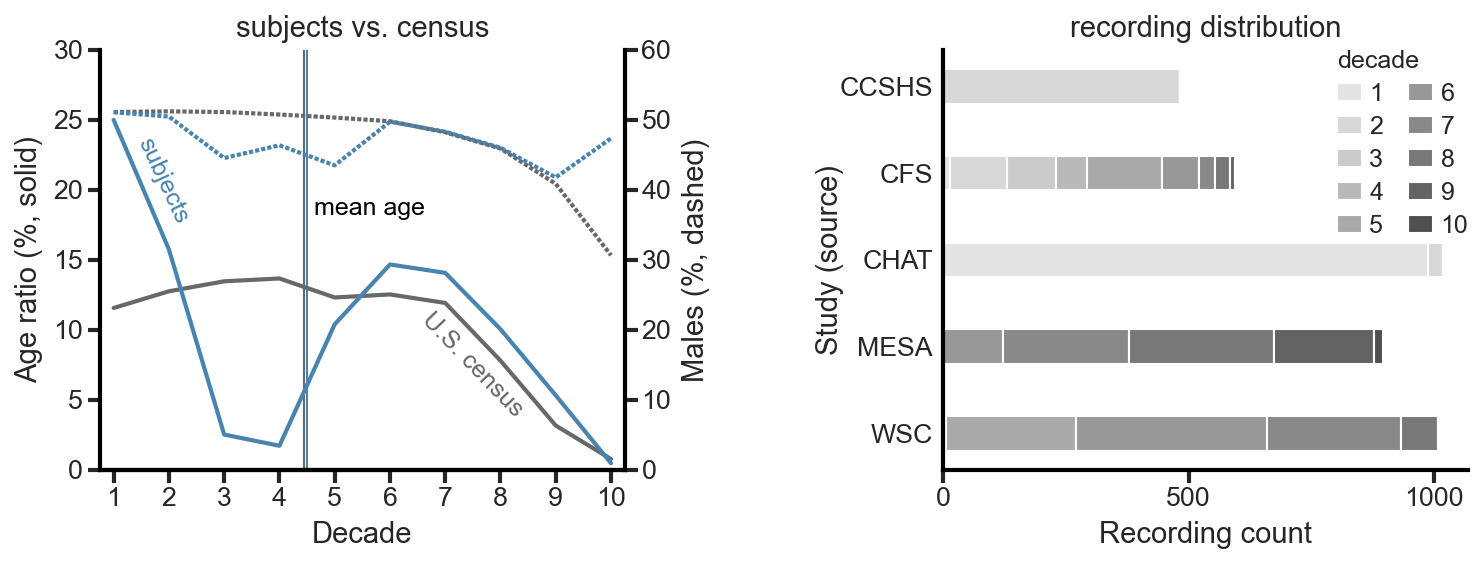
In addition to achieving expert-level performance, another primary goal was to create the most generalizable model possible. This required training and testing on a diverse pool of subjects, with the current U.S. census as the target. To meet that requirement, we used recordings from five large sleep studies (CCSHS, CFS, CHAT, MESA, WSC; all from NSRR) to build the desired dataset.
Methodological rigor
We took significant measures to address the common methodological shortcomings in the literature. Here are a few bullets on the rigor:
- We took all recordings as-is and did not trim wake periods before the subject fell asleep (mean sleep latency = 1.3 ± 1 h) or after the subject woke up.
- 4000 recordings were randomly selected from the larger pool of 5718 recordings that met our quality criteria. Those recordings were then randomly assigned to a set, with the target of all three sets having matching distributions of age, sex, and source study.
- Testing set: 500 unique recordings from unique individuals. No individuals from the validation or training set.
- Validation set: 500 recordings, but more than one recording is allowed from the same individual. No individuals from the testing or training set.
- Training set: 3000 recordings, but more than one recording is allowed from the same individual. No individuals from the testing or validation set.
Available Models
There are currently three models in the repository that anyone can use to score their own ECG data:
Paper models
- Primary model
- This model is designed to take an entire night’s worth of sleep and score it all at once.
- Real-time model
- This model is designed to take just the data that has been recorded up till “now” and score it all at once. As each new 30-second epoch of data is recorded, the model can be run again.
Post-paper models
The paper’s publication has led to ongoing discussions with others. As a result, a few models have been trained since the paper was released to address some concerns that have been raised. These models have a slight (<1%) performance impact, as measured with Cohen’s kappa. However, the differences are not statistically significant (i.e., the models are equivalent in performance).
- Primary model without demographics
- The model works the same way as the primary model, except you do not need to provide the demographics (age and sex) of the subject.
- Primary model without time
- You do not need to provide the midnight offset.
- Primary model only ECG
- You only need to provide the ECG input.
- You only need to provide the ECG input.
History and Plans
You can read more about the history and future plans here.
References
Our paper
If you find this repository helpful, please cite the paper:
- [1] Adam M. Jones, Laurent Itti, Bhavin R. Sheth, “Expert-level sleep staging using an electrocardiography-only feed-forward neural network,” Computers in Biology and Medicine, 2024, doi: 10.1016/j.compbiomed.2024.108545.
- Now that the embargo period has passed, I can share the accepted manuscript version.
- Contact details provided on this website and inside the paper.
- Preprint version of the paper @ medRxiv
- Unfortunately, medRxiv does not allow updating the preprint with the accepted manuscript. While there were some large changes between the preprint and the final, published, version (mainly revolving around adding a large meta-analysis), the network and testing set results are the same.
Additional works cited
- [2] T. Wang, J. Yang, Y. Song, F. Pang, X. Guo, and Y. Luo, “Interactions of central and autonomic nervous systems in patients with sleep apnea–hypopnea syndrome during sleep,” Sleep Breath, vol. 26, pp. 621–631, 2022, doi: 10.1007/s11325-021-02429-6.
- [3] T. Penzel, J. McNames, P. de Chazal, B. Raymond, A. Murray, and G. Moody, “Systematic comparison of different algorithms for apnoea detection based on electrocardiogram recordings,” Medical and Biological Engineering and Computing, vol. 40, no. 4, pp. 402–407, Jul. 2002, doi: 10.1007/bf02345072.
- [4] H. Hilmisson, N. Lange, and S. P. Duntley, “Sleep apnea detection: accuracy of using automated ECG analysis compared to manually scored polysomnography (apnea hypopnea index),” Sleep Breath, vol. 23, no. 1, pp. 125–133, Mar. 2019, doi: 10.1007/s11325-018-1672-0.
Readers and Mentions
Expert-level sleep staging using an electrocardiography-only feed-forward neural network